Developability comes for free...?
Did AI de novo antibody generation models learn developability properties without explicitly being trained on this?
“I won’t name names of other AI biotechs — you know them very well — but I feel like they always talk about, ‘We just need more data,’ and ‘We don’t have enough data.’ It feels like a bit of a crutch. Like, make your algorithms better, your models better. You do have enough data — if you were innovative enough on your algorithm side.” - Demis Hassabis (link)
Disclaimer: this is more of vibes piece. If people are interested, I might write this up into a paper.
Last year was a tremendous year for de novo protein design. Academic groups, startups, and even chinese tech companies for some reason (?) developed new models and workflows that successfully generate de novo minibinders, VHH, scFvs, and/or mAb.
Surprisingly, it wasn’t a single convergent technique that yielded these outcomes, but rather a couple of different ones that each report decent binders at a single or double digit hit rate against a variety of targets.
Other people have compared the different approaches like here, and new ones are popping up everyday like AbSci’s Origin-1 just last week. The point is, this is not just hype. These models are real and will actively change how drugs are made very soon!
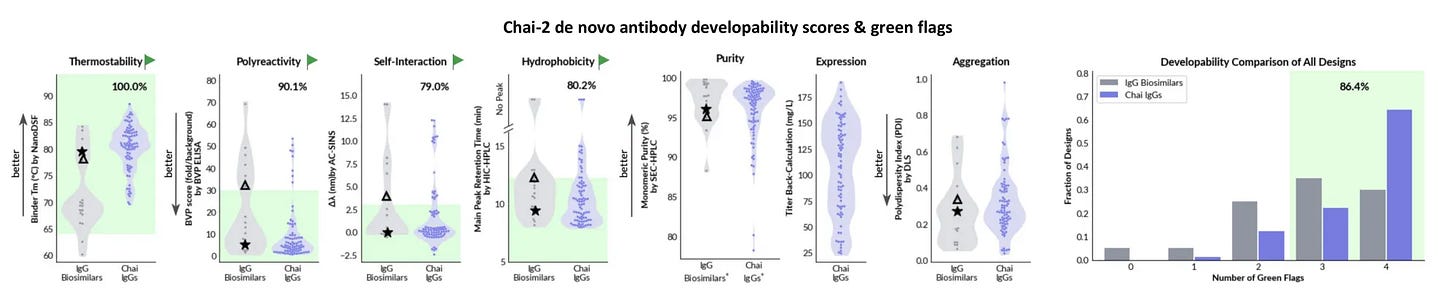
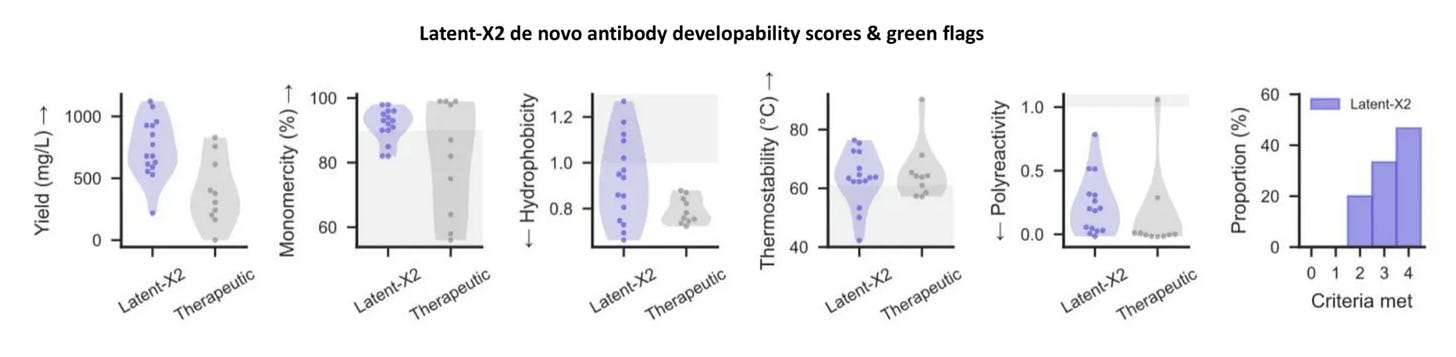
Out of the slew of new methods, one thing really caught my eye from Chai’s and Latent Labs’ most recent publication. Both Chai-2 and Latent-X2 generated antibodies that didn’t just bind well - they looked like drugs! They expressed well, were stable, didn’t aggregate, and showed limited binding to other targets.
These sort of developability characteristics can be quite a nuisance to optimize for, taking several months of work. Numerous CROs and startups have made this sort of multi-objective optimization their main selling point.
It’s worth noting that neither Chai nor Latent is the first to do this. Late in 2024, Nabla showed that JAM-1 could generate de novo VHHs that could meet “standard industry” metrics for antibody developability, and their more recent JAM-2 arguably achieved better performance than Latent X-2 and Chai-2. So why make so much fuss about these new models?
Latent made a bold proclamation:
[Our] designed molecules exhibit developability profiles that match or exceed those of approved antibody therapeutics, including expression yield, aggregation propensity, polyreactivity, hydrophobicity, and thermal stability, without optimization, filtering, or selection. […] Representative de novo VHH binders targeting TNFL9 exhibit both potent target engagement and low immunogenicity […] These properties emerge directly from the model, demonstrating the therapeutic viability of zero-shot molecular design.
They claim they didn’t train or filter on any supervised dataset for developability, but their antibodies had these properties. From the looks of it, Chai-2 may not have been trained on any developability data either. This is mostly speculative and based on their team size and composition and no reference to any custom datasets for this task. Perhaps they used some small public datasets as part of their scoring metrics; this is unclear.
For context, Nabla ran over 70 de novo antibody campaigns1 over the course of multiple years to generate rich datasets to train and align their models on. By all common wisdom, this sort of data moat is exactly what you would want. Chai and Latent did none of this and seemed to attain competitive results.
So is developability just an “emergent property” of de novo models? Does developability come for free?
But why? how? what???
There is no free lunches, right? If the models are good at this task, then there must be some reason. So either 1) the metrics that people are using are actually just kinda bad, 2) developability is encoded in/learnable from the training data somehow, or 3) some secret third thing?
Are the metrics real?
For some games, the easiest way to win is to cheat. There is a bit of history of people using creative definitions to claim victory in de novo antibody design, so yea, it’s worthwhile to double checking that the metrics are real.
So how is developability defined?
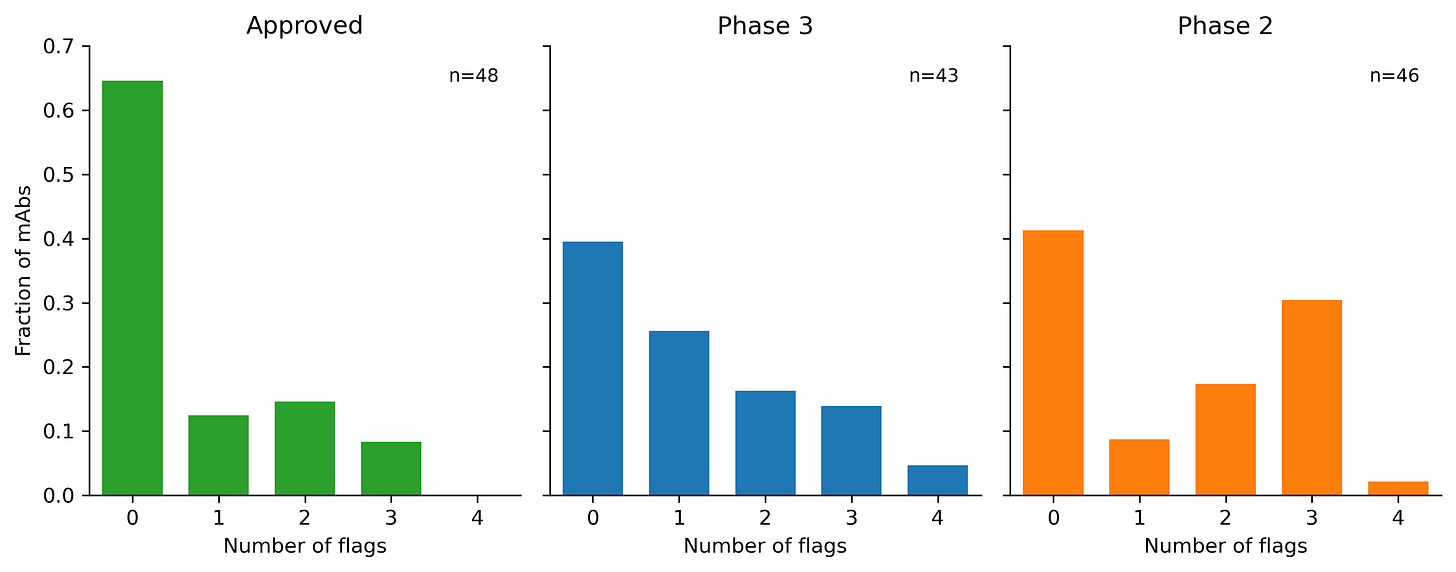
Basically, everyone points back to Jain 2017 paper, which does a great systematic assessment of nearly all the clinical stage antibodies (CST) at the time. Jain et al. reformat the antibodies to all be IgG1 and then ran 12 biophysical assays on them. They cluster the assays into 5 groups, but I think there should be at least 6: expression (HEK titer), thermostability (Tm), hydrophobic interaction (SGAC100, SMAC, HIC), long-term aggregation propensity (AS), antigen promiscuity (ELISA, BVP), and cross-interaction/self-interaction (PSR, CSI, AC-SINS, CIC). If you wanna know more about these assays, then read this. Jain et. al basically just calls the bottom 10% from each group of properties (not including expression or thermostability) as a red flag, then counts the number of flags.
The paper was more making heuristics rather than trying to generate super rigorous classifier thresholds. There are too many confounders in this approach to really make such a classifier, but you can see pretty clear trend lines. The later stage CST have fewer flags on average. Since people didn’t really start running these assays until after all of these CST were already made, we probably aren’t getting Goodharted.
Yes, sometimes drug dev is more an art than a science, see Lipinski rule of five.2
Chai and Latent both try to reproduce a few of these assays using a subset of the CST antibodies. They seem to do a decent job. After adjusting for some scaling factors, they generate their own set of flag thresholds.
They both cover a decent range of developability properties. The exact explanation for their thresholds is a bit murky, but overall, this all seems reasonable. I tried to compare them using reference antibodies, and Latent seems to have looser thresholds. In particular, the Lirilumab HIC retention threshold is hard to validate.3
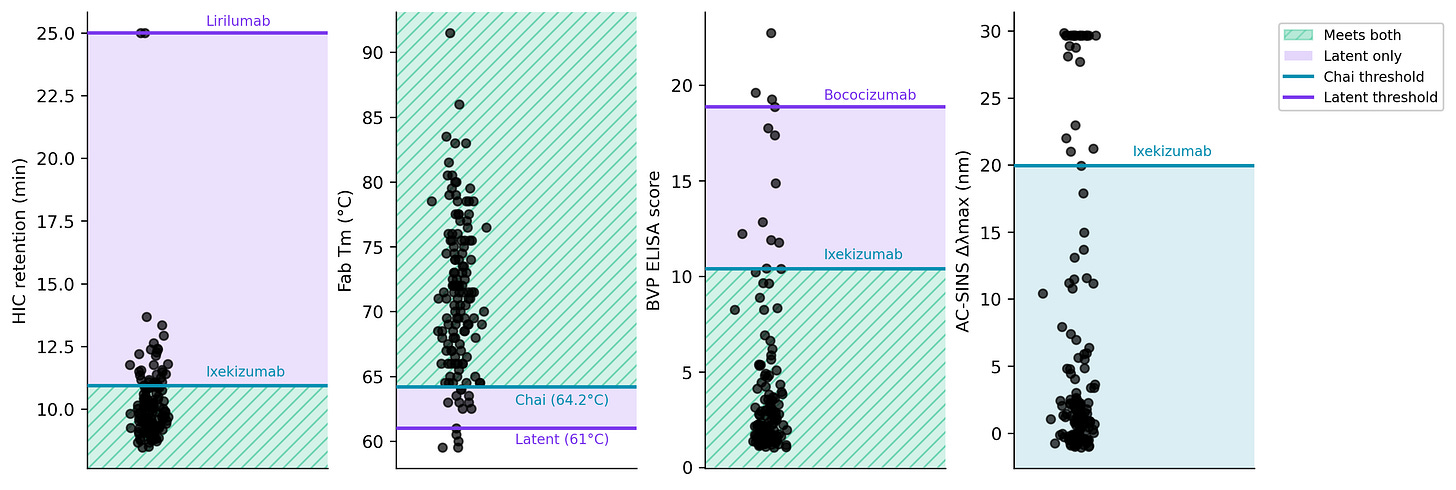
On these thresholds, Chai and Latent report >85% and >80% of their designs having 3 or 4 green flags out of 4, respectively. 👀
So yea, the assays are real, their thresholds seem pretty reasonable, and honestly, >80% passing from zero-shot is damn good. How are they doing this?
Maybe it’s all in the training data?
Put simply: models can only learn from what is in their training data. If they are generalizing out of distribution, then somehow they are learning this from the training data.
There has been much ink spilled on whether or not AlphaFold and other AI protein models are really learning physics or not. It is a pretty big claim and would require a commensurate level of proof. I think something simpler is happening. The training data just has nice structures. Really nice structures. Nicer than people (or at least I) appreciated.
If you want to structurally characterize a protein, you first need to express the protein. The expressed protein (and any binding partner) needs to be decently stable to withstand purification, freezing, and whatever else you do to it. To resolve a high quality structure, you also need the protein to not aggregate too much. And of course, you need to convince someone to give you money to run this experiment and care enough to run it. So the protein is likely “important” in some way4.
Thats a lot of selection bias!
There aren’t really that many antibody structures in the PDB (~5-10k), and only a small number of them are CST/approved (~200). It isn’t clear that the majority of these structures have been optimized to have great developability properties. Is this selection bias all you need?
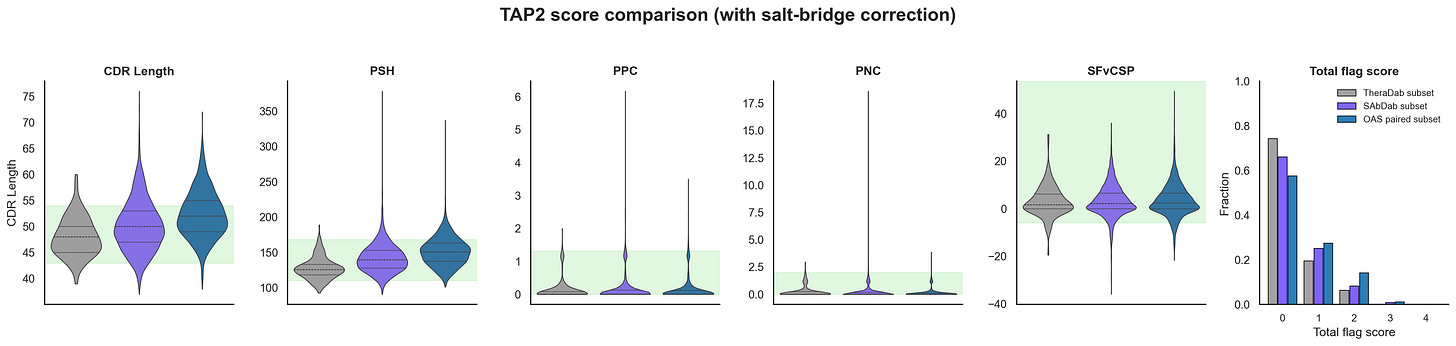
One way to try to suss this out is to just score all the antibody structures in the PDB with in-silico developability tests and see if they are really good. Raybould et al made the Therapeutic Antibody Profiler 2 (TAP2), which tries to capture Jain 2017 vibes metrics with in-silico alternatives and shows that the total flags generate similar trend lines. I found a bug in the salt-bridge correction while reproducing TAP2 that changes the total flags. I reached out to Raybould and OPIG, and they are fixing it now. In the meantime, I will include both scores, but it mostly doesn’t change the results too much.
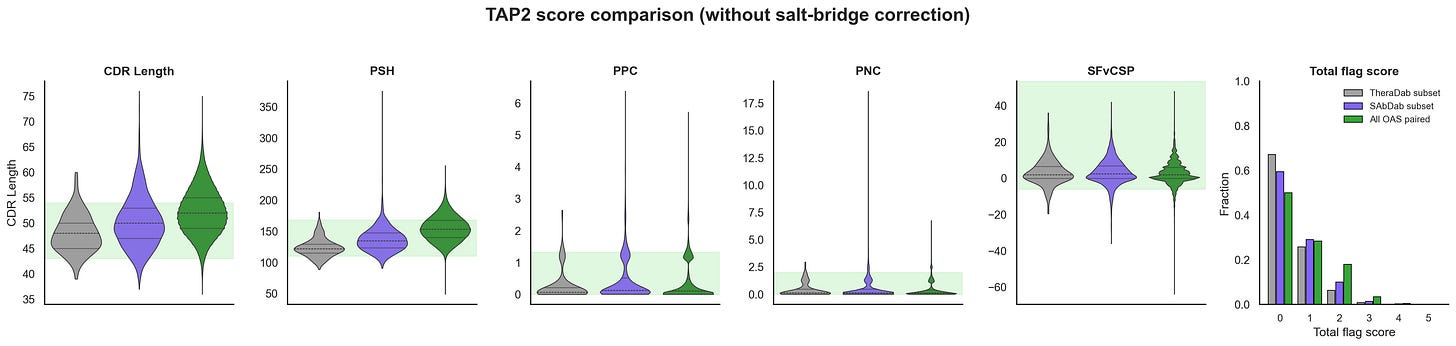
Based on the TAP scores, the antibody structures in PDB (i.e. SAbDab) actually look like they are quite developable and better than natural sequences from OAS paired (i.e. statistically significant). Even when you remove the CST antibody structures from PDB and compare with OAS naive and memory/plasma B-cells separately, you still see this bias in SAbDab (plots not shown). Most of this difference seems to be coming from CDR Length and PSH, which are decently correlated. That said, OAS paired sequences already have decent developability scores. This isn’t a super rigorous analysis, but seems like something… cool!
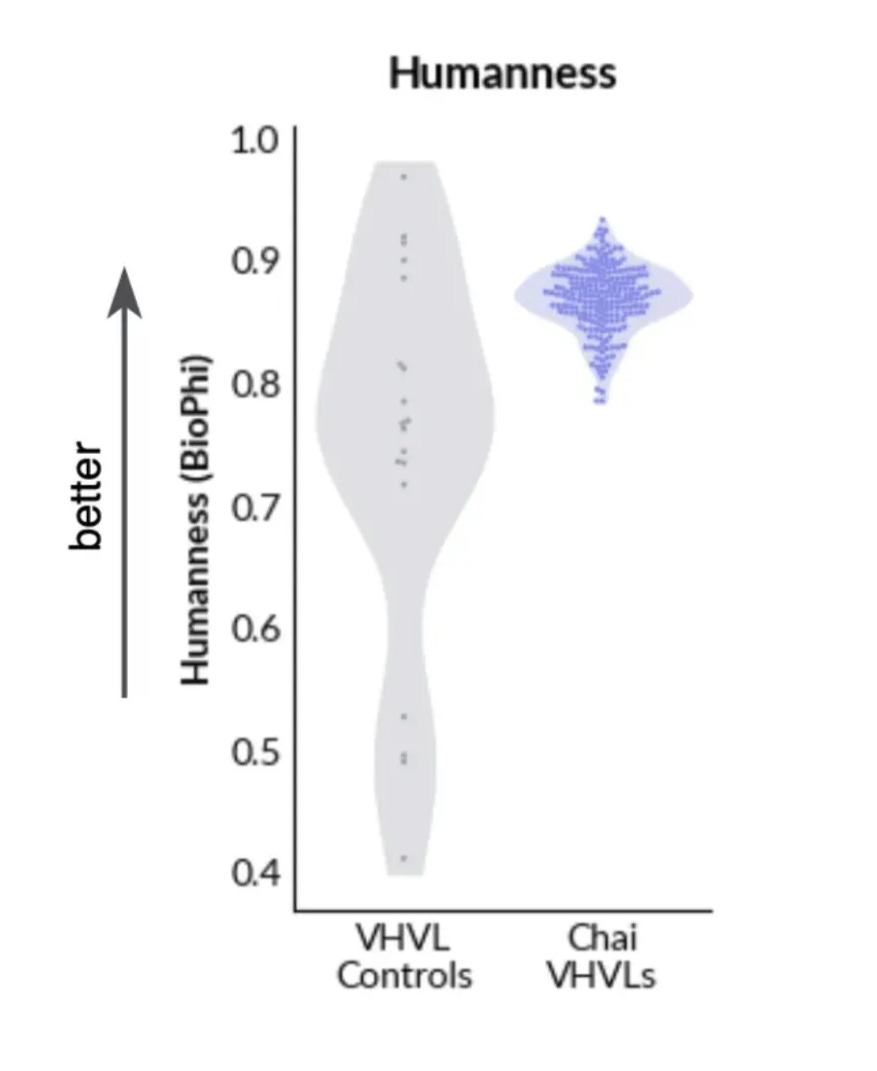
For immunogenicity, Chai doesn’t do any experimental assessment; instead, they do an in-silico assessment for the “humanness” of the antibody sequence using BioPhi/promb package. The de novo sequences look pretty good!
I did a similar analysis for the PDB. As expected, the OAS sequences and CST human antibodies show up as very human. But the antibody structures in the PDB are on average as human as a CST humanized antibody.
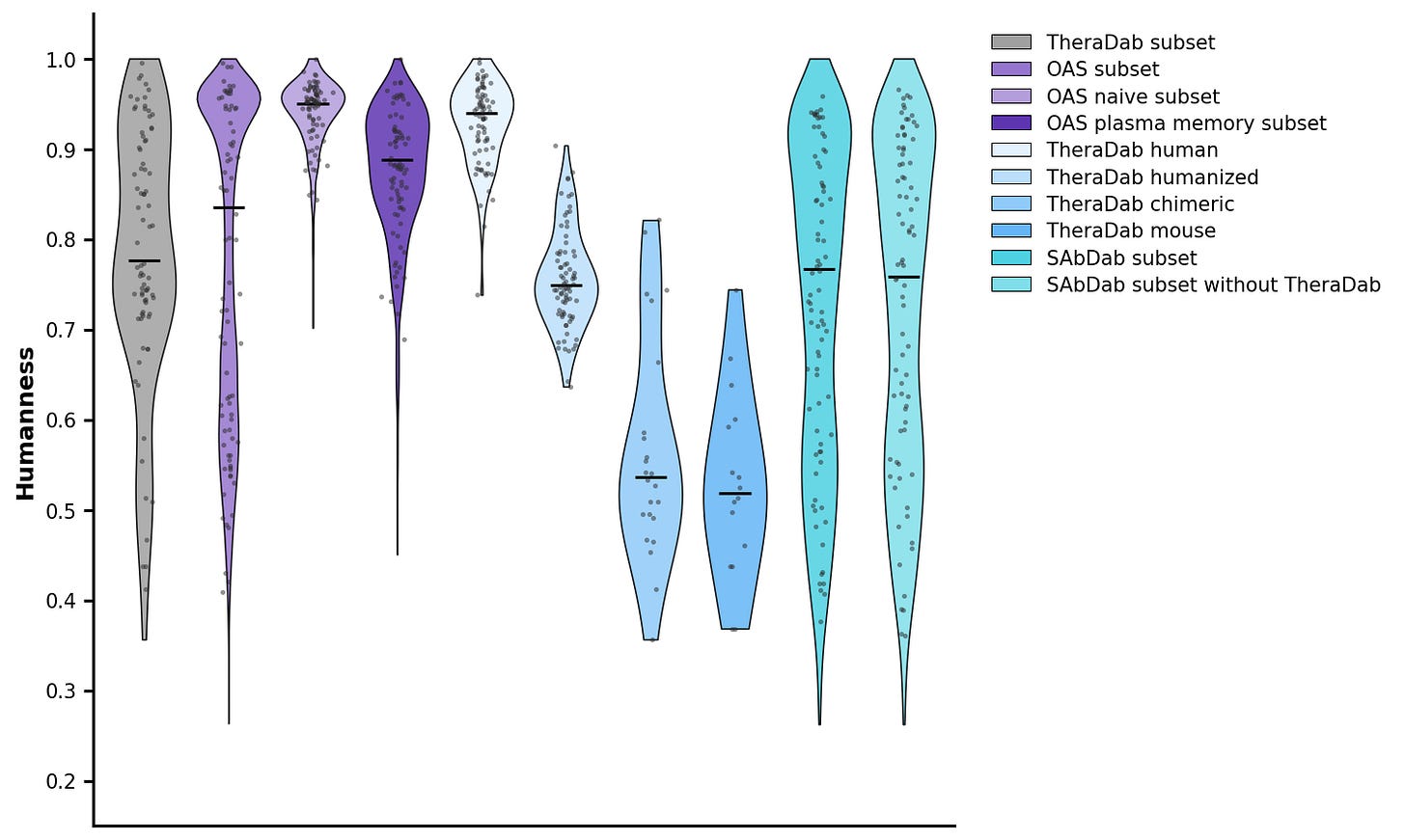
At least on these in-silico metrics, the antibodies in the PDB definitely look kinda like drugs!
Latent actually experimentally tested immunogenicity for a handful of their de novo VHH constructs on primary immune cells isolated from 10 healthy donors. They used T-cell proliferation and cytokine release assays and show no change for any of their VHHs for any of the donors! Perhaps Chai antibodies will also be non-immunogenic…
Does the bias of antibody structures in the PDB towards good developability properties explain the emergent properties of these models? Perhaps synthetic structures from OAS are good enough?
Some secret third thing?
We don’t really know how the sausage got made. So perhaps there is some secret third thing that we don’t know about that is making their antibodies developable.
It is not clear that using “good” framework sequences would have such an effect since some developability properties are not even framework specific (e.g. aggregation, polyreactivity). Chai notes that they are using VH3-23 and VH3-66 which have good biophysically properties in Jain 2017. Similarly, Latent uses 5 validated framework sequences when generating their de novo antibodies. If that was all you needed to do, then everyone would just use these amazing frameworks and rarely need to do much optimization. Also, Chai-2 antibodies seem to outperform framework-matched controls on at least thermostability. There are more nuanced counterarguments to be made, but I couldn’t come up with anything convincing.
Nabla claims that they used over 70 de novo antibody campaigns to train JAM. Did Chai or Latent secretly generate or acquire a huge developability dataset? Can you use small developability datasets and in-silico metrics to effectively align de novo models or score de novo antibodies? Is there some special property of synthetic data generation or new training/inference technique they figured out? Something else??
On the bright-side, if you don’t really need any special data, then we should be quite optimistic about open-source models. Perhaps Boltzgen antibodies (or some other open-source model) have good developability properties already!
Conclusion
So, on these pretty reasonable developability metrics/thresholds, Chai and Latent both generate de novo antibodies have “good” developability. They seem to do this without training, filtering, or selecting using developability data, or at least not much of it.
There is potentially some developability bias in the structure data that these models were able to learn as they scaled, or maybe there is also some special sauce that isn’t being disclosed.
From my best guess, some mixture of thoughtful selection of nice epitopes, training data biases, scoring for binding, filtering for liabilities (and developability?), and use of good frameworks was sufficient to nudge us into a really good regime.
Perhaps Demis is right. Maybe we don’t need more data. Maybe we just need better algorithms, better models. Can PK/PD be learned from the structure data we already have [link, link]? What else are we leaving on the table? One can only wonder what Isomorphic is up to…
We are getting close to the day where antibody discovery and optimization is all done on GPUs. It just feels like we skipped a step to get there, and I think this is worth appreciating. Can’t say for sure what is happening, but it would sure be nice if developability came for free.
Thanks to AM, BN, DN, HK, and VH for giving me feedback! I hope to write more this year, so stay tuned…
not all of these include developability metrics
developability is a vague catch-all term that covers all non-therapeutics properties that can be developed; at least Lipsinki’s rule of five is a concrete list with explicit ranges.
Nabla has different thresholds…
all proteins are important 😁


My impression from reading what Nabla has disclosed is not that they ran those many campaigns in order to obtain developability data. Rather, it seems they used those campaigns to tune their method and its hyperparameters. (For example, the Nipah contest results suggest that ipSAE is better than ipTM for selecting designs; this is the sort of thing one can figure out from running many campaigns.) Of course, their method and hyperparameters may have been tuned to achieve good developability. But I think it's unlikely that they've trained universal developability oracles. Such oracles are useful if you're operating on the edge or beyond the edge of developability, and need to move to the safe side of the boundary. But, as you say, it may be possible in some cases to instead simply avoid the danger zone.